ERC Advanced Grant "SoilLife"
Period: 2013-2018, ERC No 320499
“The Hidden Frontier: Quantitative Exploration of Physical and Ecological Origins of Microbial Diversity in Soil”
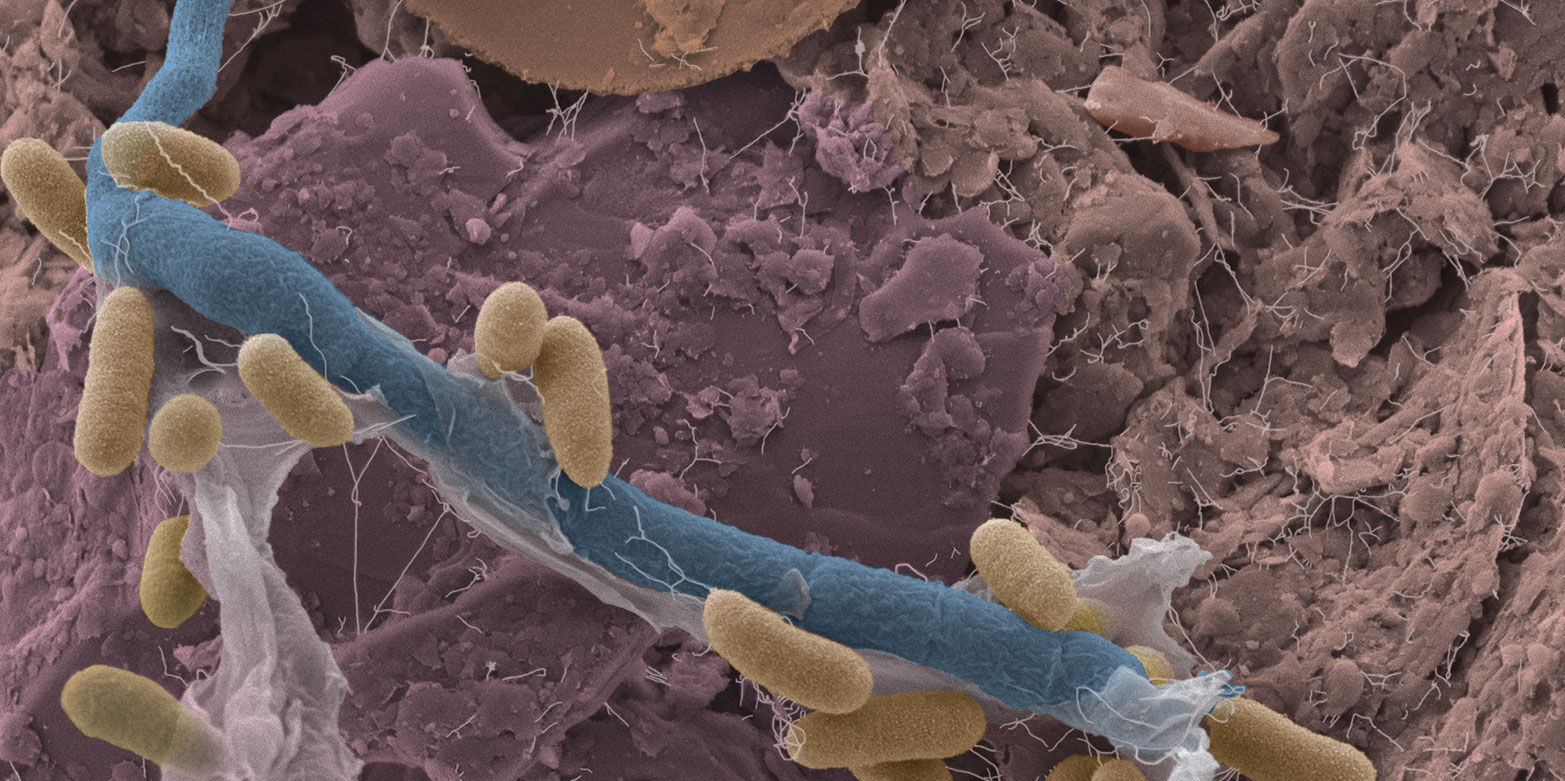
By some accounts exploring the origins of soil microbial diversity represents a scientific frontier similar to that of space exploration in its scope. This biodiversity is attributed to soil ecological heterogeneity reflecting interplay of physical, chemical and other variables that define highly dynamic microbial habitats and function. This ERC project aims to quantify the role of the physical environment, defined by pore-space architecture as well as presence and organization of soil water, in shaping microbial populations and interactions and thus maintaining the unparalleled diversity and ecological function of the soil, which is arguably the biologically richest compartment of the earth’s biosphere.
The researchers will develop a novel individual-based modeling platform that simulates microbial life within soil pores, explicitly considering the dynamic aqueous phase network that supports a host of biophysical processes (e.g., motion and dispersion of organisms, nutrient diffusion, etc.). The resulting virtual soil microcosm will enable systematic exploration of the hidden microbial life under different environmental and climatic scenarios enabling quantitative prediction of how soil biophysical processes affect water quality, the fate of pollutants in soil, and the functioning of global biogeochemical cycles.
SoilLife researchers:
Principal Investigator:
Prof. Dani Or
Members:
Sami Ben Said, Samuel Bickel,
Benedict Borer, Minsu Kim, Hannah Kleyer, Dr. Robin Tecon
Former members:
Dr. Gang Wang, Dr. Olga Ilie, Dr. Ali Ebrahimi
Recent publications from SoilLife
- Spatial organization of bacterial populations in response to oxygen and carbon counter-gradients in pore networks. Benedict Borer, Robin Tecon and Dani Or. Nature Communications, 9:769. DOI
- Dynamics of soil biogeochemical gas emissions shaped by remolded aggregate sizes and carbon configurations under hydration cycles. Ali Ebrahimi and Dani Or. Global Change Biology, 24 (1): e378-e392, 2018. external page DOI
- Resolving species level changes in a representative soil bacterial community using microfluidic quantitative PCR. Hannah Kleyer, Robin Tecon and Dani Or. Frontiers in Microbiology, 8: 2017, 2017.external page DOI
- Biophysical processes supporting the diversity of microbial life in soil. Robin Tecon and Dani Or. FEMS Microbiology Reviews, 41 (5): 599-623, 2017. external page DOI
- Synthetic Microbial Ecology: Engineering Habitats for Modular Consortia. Sami Ben Said and Dani Or. Frontiers in Microbiology, 8: 1125, 2017. external page DOI
- Mechanistic modeling of microbial interactions at pore to profile scale resolve methane emission dynamics from permafrost soil. Ali Ebrahimi and Dani Or. Journal of Geophysical Research. Biogeosciences, 122 (5): 1216-1238, 2017. external page DOI
- Microbial community response to hydration-desiccation cycles in desert soil. Adam Štovíček, Minsu Kim, Dani Or and Osnat Gillor. Scientific Reports, 7: 45735, 2017. external page DOI
- Cooperation in carbon source degradation shapes spatial self-organization of microbial consortia on hydrated surfaces. Robin Tecon and Dani Or. Scientific Reports, 7: 43726, 2017. external page DOI
- Hydration status and diurnal trophic interactions shape microbial community function in desert biocrusts. Minsu Kim and Dani Or. Biogeosciences, 14 (23): 5403-5424, 2017. external page DOI
Recent research activities:
Microgeography of microbial life in soil (Samuel Bickel)
In order to better understand the characteristics and spatial distributions of microbial settlements in soil pore spaces, we study and model the cells’ microenvironment trough considerations of soil properties, resource fluxes and climatic conditions. The distribution of resources in the soil profile shapes the abundance of microbial biomass. By considering physical soil structure and climatic conditions, we can identify zones of high microbial activity at the profile scale
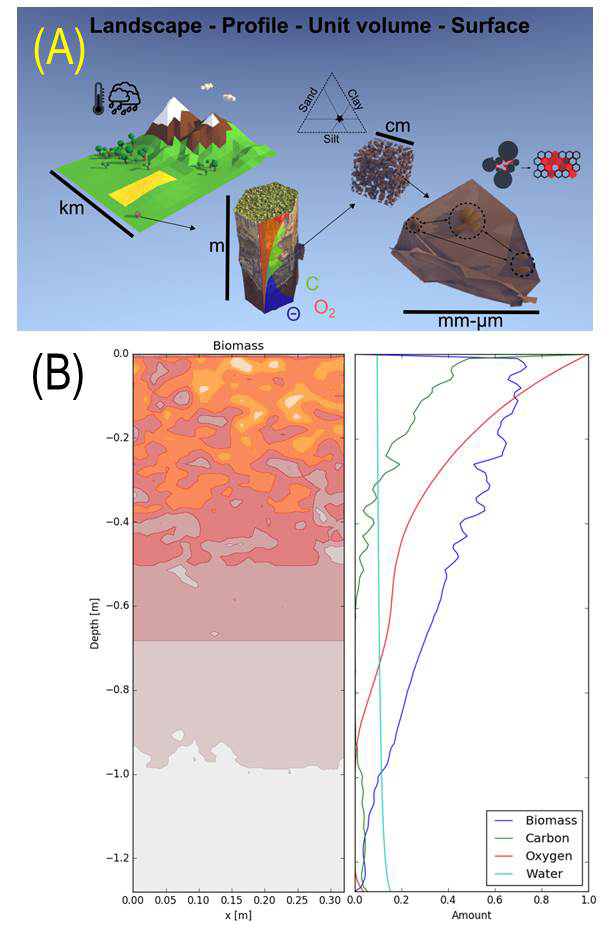
Only a small fraction of the total soil surface area is colonized by microorganisms. We study this microbial spatial distribution experimentally using a Porous Surface Model (PSM) as an analog of soil pore surfaces. In parallel to the experimental approach, we develop an individual-based model that considers cell shoving to estimate the size of microbial settlements
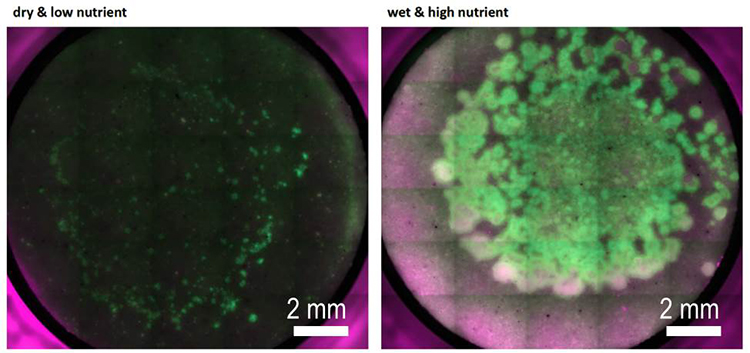
Species-level qPCR analysis of soil bacterial assembly under changing environmental conditions (Hannah Kleyer)
We have assembled a synthetic community of traceable bacterial species commonly found in natural soils, which allows us to systematically study community dynamics in response to abiotic factors (e.g., soil matric potential). The synthetic community is incubated in sand microcosms under controlled, fluctuating hydration and nutrient conditions; microorganisms grow and interact in this spatially complex pore network, and the community composition is assessed at the species level using genetic fingerprinting. We are particularly interested in how hydration dynamics affect composition and diversity of the synthetic soil microbial assembly in unsaturated microcosms

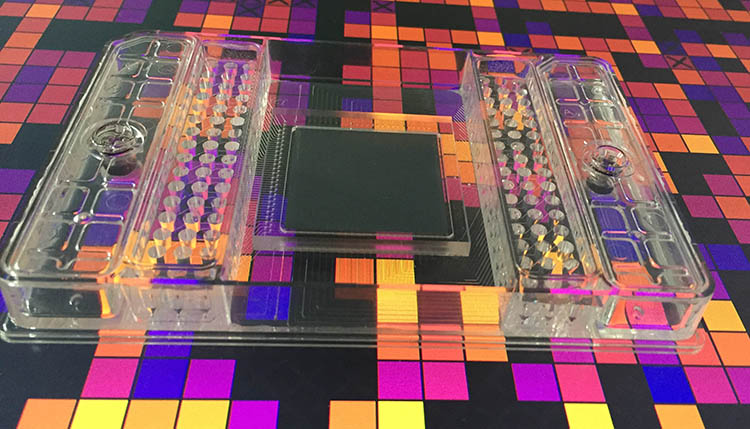
We use quantitative PCR at the species level to analyze the community composition in our microcosms. A new protocol was established in our lab for high-throughput quantification of bacterial community composition using a microfluidic reactor technology available at the Genetic Diversity Center of ETH Zürich (http://www.gdc.ethz.ch)
Read more about the new method in the original article:
Resolving species level changes in a representative soil bacterial community using microfluidic quantitative PCR. Hannah Kleyer, Robin Tecon and Dani Or. Frontiers in Microbiology, 8: 2017, 2017.external page DOI
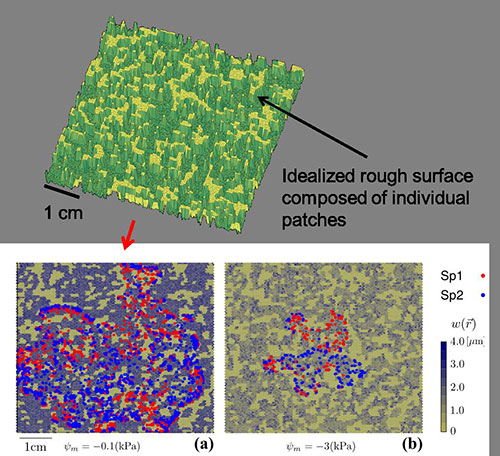
Modeling microbial life on hydrated rough soil surfaces (Minsu Kim)
We have developed a Rough Surface Patch model (RSPM) to study microbial life in soil. In this model, the key element of water retention is encapsulated in patch roughness properties that collectively present soil water retention properties and aqueous phase distribution at a given matric potential. The scalable representation of surfaces as patches enables simple computations for ecological modelling at sub-metric scales and for days to months time frames. Larger simulation domain permits considerations of temperature and hydration gradients known to affect microbial ecology near interfaces (i.e., soil surface, desert biological soil crust etc.).
Recently, we have used RSPM to explain and predict the spatial self-organization and activity of microbial communities in desert biocrusts:
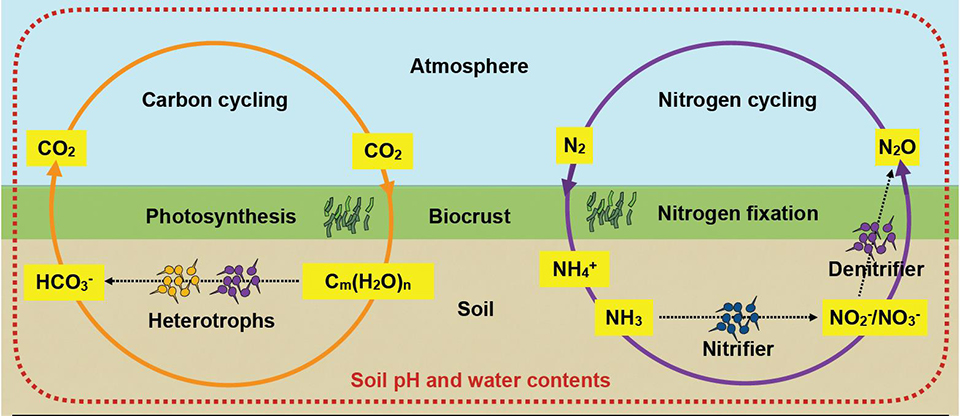
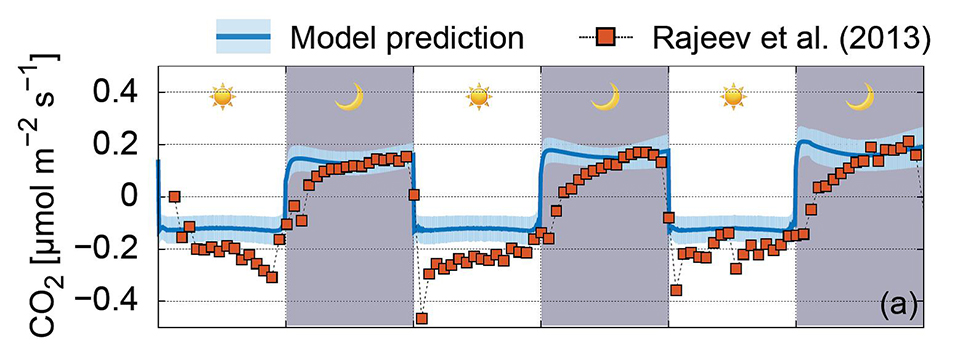
Read the original article:
Hydration status and diurnal trophic interactions shape microbial community function in desert biocrusts. Minsu Kim and Dani Or. Biogeosciences, 14 (23): 5403-5424, 2017. external page DOI
The RSPM has also been used to explain the response of microbial communities to drying-wetting cycles in desert soil, based on direct comparisons with field observations. Read the original article:
Microbial community response to hydration-desiccation cycles in desert soil. Adam Štovíček, Minsu Kim, Dani Or and Osnat Gillor. Scientific Reports, 7: 45735, 2017. external page DOI
Read about the RSPM in the original article:
Individual-Based Model of Microbial Life on Hydrated Rough Soil Surfaces. Minsu Kim and Dani Or. PloS ONE (2016). external page DOI
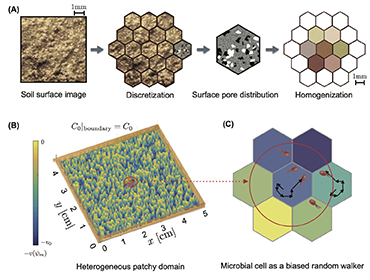
Activity of microbial communities in soil aggregates (Ali Ebrahimi)
Using a 3-D pore network that integrates individual-based models (IBMs), we systematically evaluate the effects of pore geometry and hydration conditions on microbial interactions and motion in unsaturated soil. The model allows us to quantify the role of the fragmentation of aqueous (micro-)habitats on bacterial dispersal rates.
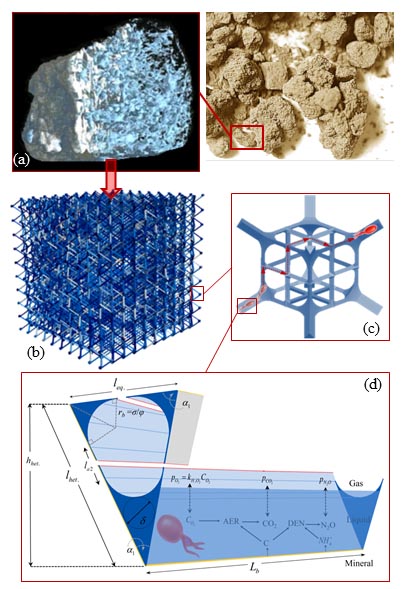
Read the original article:
Microbial community dynamics in soil aggregates shape biogeochemical gas fluxes from soil profiles – upscaling an aggregate biophysical model. Ali Ebrahimi and Dani Or. Global Change Biology, 22 (9): 3141-3156, 2016. external page DOI
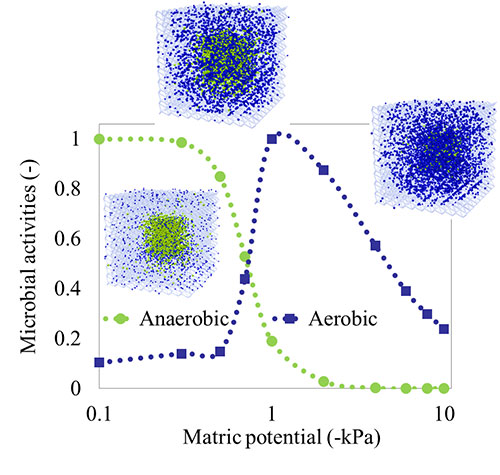
The model is used to quantify the effects of hydration states on oxygen diffusion and on microbial aerobic/anaerobic activity in soil aggregates. The model offers new insights into the dynamics of microbial communities that shape key soil biogeochemical processes such as respiration and denitrification.
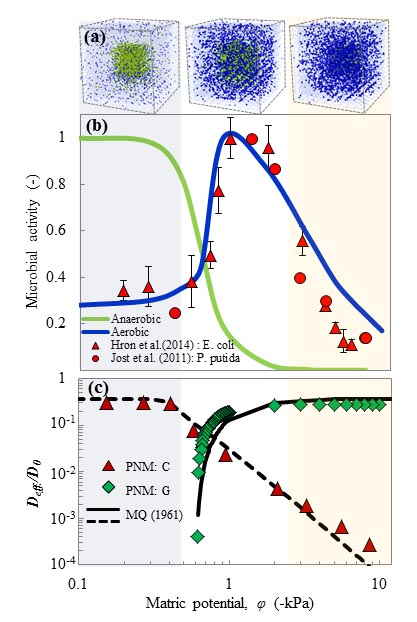
Read the original article:
Hydration and diffusion processes shape microbial community organization and function in model soil aggregates. Ali Ebrahimi and Dani Or. Water Resources Research (2015). external page DOI
Microbial motility on porous rough surfaces (Robin Tecon)
Most soil habitats are not water-saturated and this has an important effect on microbial motility and dispersal. Using a Porous Surface Model (PSM) that mimics unsaturated soil surfaces, we investigate the flagellar motility of soil bacteria at various hydration conditions. We use 3-D scanning laser microscopy and epifluorescence microscopy to measure aqueous films and to examine bacterial movement at the microscale.
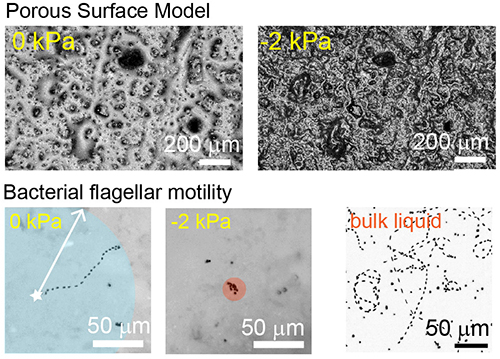
Read the original article:
Bacterial flagellar motility on hydrated rough surfaces controlled by aqueous film thickness and connectedness. Robin Tecon and Dani Or. Scientific Reports, 6 (2016). external page DOI
Engineering modular microbial consortia (Sami Ben Said)
We explore new ideas and concepts to assemble the members of a microbial community in separate interconnected bioreactors, hence allowing them to interact/communicate in a controlled way. This separation of processes aims to provide each microbial species with a more suitable set of environmental conditions (pH, temperature, oxygen and nutrients concentrations,…) than they would experience if they were incubated as a co-culture in a single bioreactor.
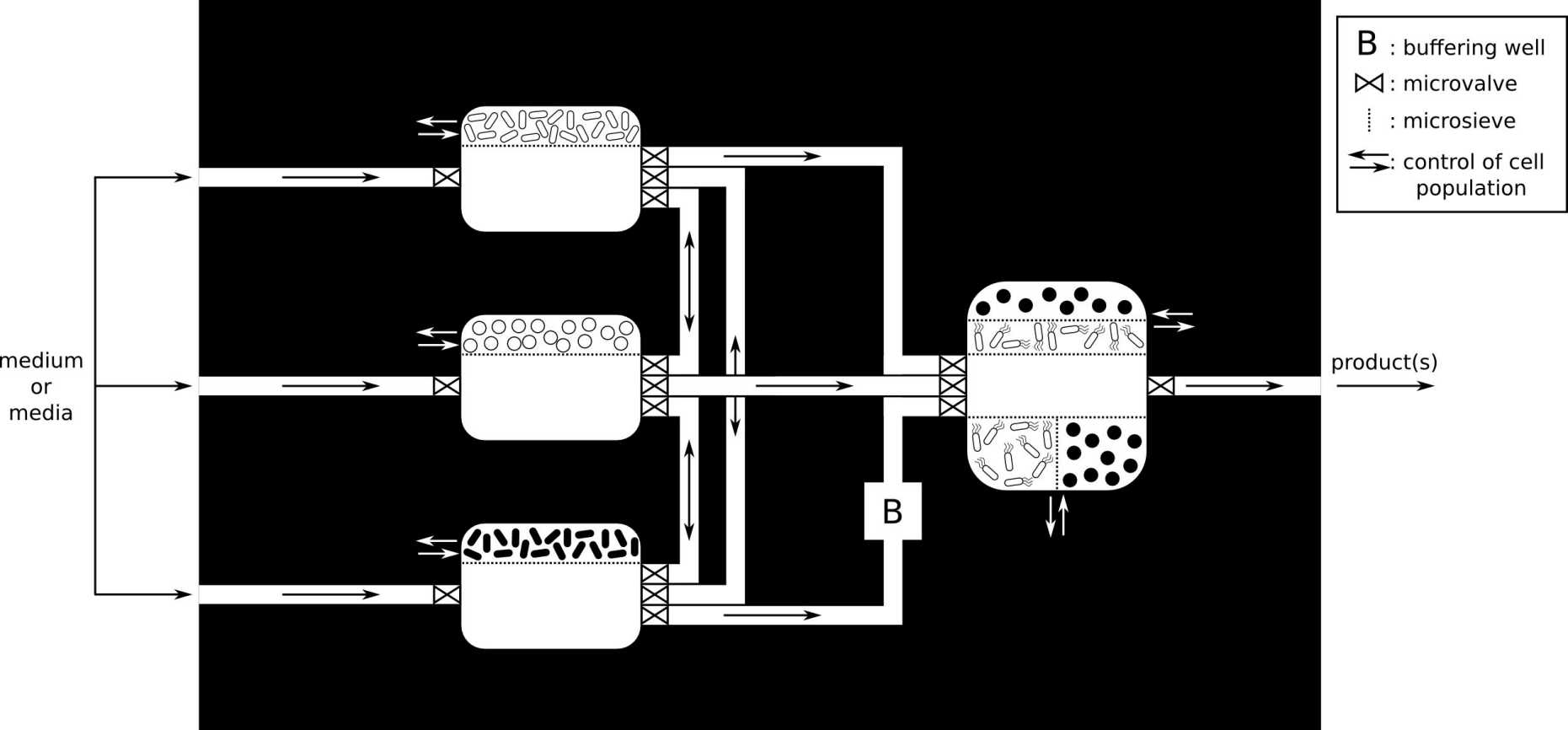
Read more about the concept of spatially linked consortia in the original paper:
Synthetic Microbial Ecology: Engineering Habitats for Modular Consortia. Sami Ben Said and Dani Or. Frontiers in Microbiology, 8: 1125, 2017. external page DOI
Mutualistic bacterial interactions (Robin Tecon)
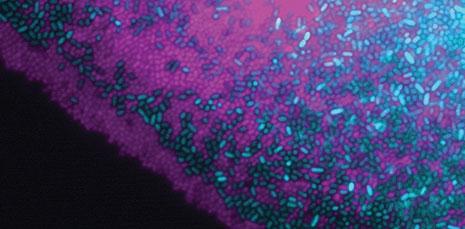
Read the original paper:
Cooperation in carbon source degradation shapes spatial self-organization of microbial consortia on hydrated surfaces. Robin Tecon and Dani Or. Scientific Reports, 7: 43726, 2017. external page DOI
On-going research activities:
Development of agent-based models to understand and predict the behavior of microbial communities in terrestrial environments
In SoilLife, we are developing various models in order to understand different aspects of the soil environment:
- The 2D Roughness Network Model (Wang & Or, 2014; Wang & Or, 2013; Wang & Or, 2010; Long & Or, 2007) is a discrete individual-based model that is spatially and temporally resolved, and we can track the motion and life history of each bacterium in the simulation. The Roughness Network model efficiently describes pore-space distribution of rough surfaces and associated aqueous-phase configuration that is determined by surface hydration conditions, and their effects on nutrient diffusion and microbial motility and growth and interactions with neighboring microbes and local environments at the microscale.
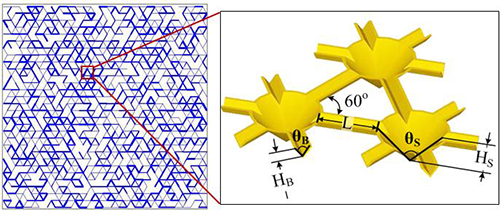
- The Soil Surface Patch Model uses discrete patches with defined porous and water properties, which helps reducing the computation power needed for simulations. This allows us to upscale from the microbial world to the macroworld (centimeter scale) and makes a link with known hydrologic models
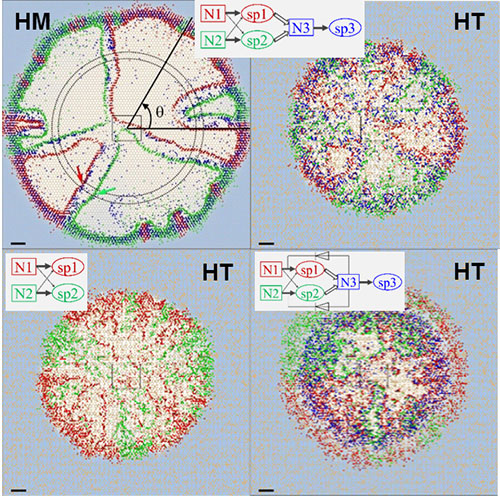
Examples of simulated results of microbial trophic interaction from soil surface patch model (SSPM). We consider two microbial species, one Red and one Blue, that grow in a wet soil (a) and in a dry soil (b). Both species use two obligatory nutrients with different affinities. Image credit: Minsu Kim
- The 3D Pore Network adds a new dimension to the Roughness Network and permits us to study the role of a 3D structure on bacterial interactions and spatial arrangements.
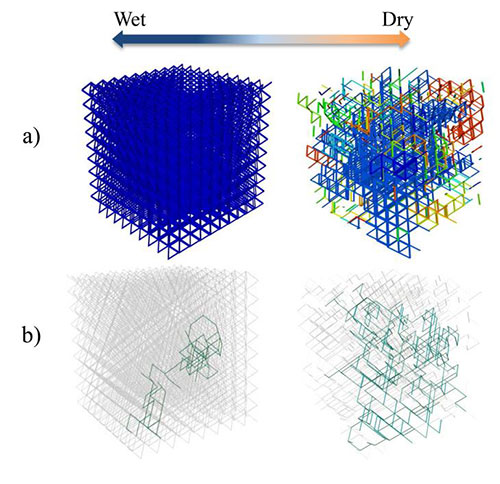
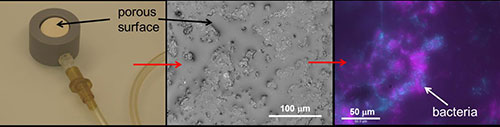
Development of experimental models to investigate the behavior of microbial communities and to test the predictions of computer simulations
We are currently using and developing several types of experimental models for the study of bacterial communities growing under saturated or unsaturated conditions.
- The Porous Surface Model, or PSM, consists of a ceramic disc covered by a water film that can be precisely controlled (Dechesne et al., 2008). The PSM is amenable to direct observations with epifluorescence and confocal microscopy, and thus we can visualize how bacteria spread on the surface under various unsaturated conditions. The PSM mimics a key parameter of soil environments, that is, the soil matric potential, which controls the availability of water to bacteria.
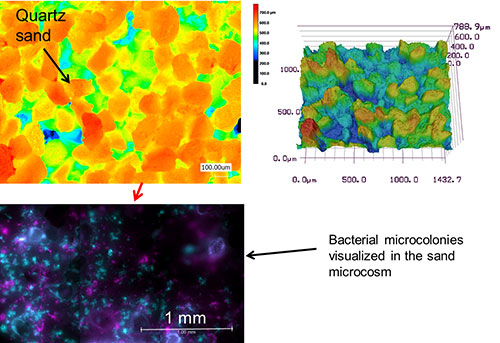
- Quartz Sand Microcosms consist of layers of fine sand particles (100-150 mm) that is spread on a PSM. This way, we can control the matric potential (i.e., water availability) in the microcosm. The sand microcosms represent a 3D variation of the regular PSM, in which microorganisms can experience a complex spatial pore network.
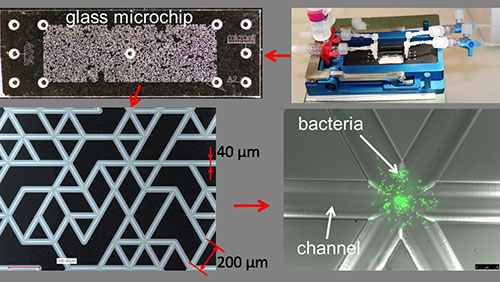
- Glass Microchips contain thousands of microchannels in order to mimic an intricate porous network such as found in terrestrial environments. These microchips can be populated with bacteria, and the glass material makes them amenable to direct observation by microscopy. We can thus observe how microorganisms grow and organize themselves in a pore network at the microscale.
List of relevant publications:
- Wang G., and D. Or, 2014, Trophic interactions induce spatial self-organization of microbial consortia on rough surfaces, Scientific Reports, 4: 6757, DOI:10.1038/srep06757
- Ebrahimi A., and D. Or, 2014, Microbial dispersal in unsaturated porous media: Characteristics of motile bacterial cell motions in unsaturated angular pore networks, Water Resources Research, 7406-7429, DOI:10.1002/2014WR015897
- Wang G., and D. Or, 2013, Hydration dynamics promote bacterial coexistence on rough surfaces, The ISME Journal, 7, 395-404, doi:10.1038/ismej.2012.115
- Wang G., and D. Or, 2012, A Hydration-Based Biophysical Index for the Onset of Soil Microbial Coexistence, Scientific Reports, 2, 881, DOI: 10.1038/srep00881
- Dechesne, A., G. Wang, G. Gülez, D. Or and B. F. Smets, 2010, Hydration controlled bacterial motility and dispersal on surfaces, The Proceedings of the National Academy of Sciences USA, 107, (32), 14369-14372, doi:10.1073/pnas.1008392107
- Wang, G., and D. Or, 2010, Aqueous films limit bacterial cell motility and colony expansion on partially saturated rough surfaces, Environmental Microbiology, 12, (5), 1363-1373, DOI: 10.1111/j.1462-2920.2010.02180.x
- Long, T., and D. Or, 2009, Dynamics of microbial growth and coexistence on variably saturated rough surfaces, Microb. Ecol., 58, 262-275
- Dechesne, A., D. Or, G. Gülez and B.F. Smets, 2008, The porous surface model: a novel experimental system for quantitative observation of microbial growth under unsaturated conditions, Appl. Env. Microb., 74, 5195-5200
- Or, D., B.F. Smets, J.M. Wraith, A. Dechesne and S.P. Friedman, 2007, Physical constraints affecting microbial habitats and activity in unsaturated porous media ? A review, Adv. Water Resour., 30, (006-7), 1505-1527
- Or, D., S. Phutane and A. Dechesne, 2007, Extracellular polymeric substances (EPS) affecting pore-scale hydrologic conditions for bacterial activity in unsaturated soils, Vadose Zone J., 6, 298-305
- Long, T., and D. Or, 2007, Microbial growth on partially saturated rough surfaces: Simulations in idealized roughness networks, Water Resour. Res., 43, W02409
Contact information
No database information available